在江蘇省中醫(yī)藥管理局科研項(xiàng)目(批準(zhǔn)號(hào):YB2020088)�、揚(yáng)州市“綠揚(yáng)金鳳”衛(wèi)生創(chuàng)新領(lǐng)軍人才基金項(xiàng)目(批準(zhǔn)號(hào): YZLYJF2020WSCX037)等資助下�����,南京鼓樓醫(yī)院集團(tuán)儀征醫(yī)院王小紅主任醫(yī)師團(tuán)隊(duì)在紅景天苷治療重癥急性胰腺炎機(jī)制方面取得進(jìn)展。研究成果以“紅景天苷通過調(diào)節(jié) miR-217-5p/YAF2 軸減輕重癥急性胰腺炎觸發(fā)的胰腺損傷和炎癥(Salidroside alleviates severe acute pancreatitis- triggered pancreatic injury and inflammation by regulating miR-217-5p/YAF2 axis)”為題��,于2022年8月10日在線發(fā)表于《國(guó)際免疫藥理學(xué)》(International Immuno- pharmacology)����。文章鏈接:https://doi.org/10.1016/j.intimp.2022.109123

急性胰腺炎(Acute pancreatitis)是臨床常見的急腹癥之一,其以輕癥多見���,病程呈自限性,其中10%~30%的患者會(huì)進(jìn)展為重癥急性胰腺炎(Severe acute pancreatitis, SAP)�����。SAP病情進(jìn)展快�,病情危重,常并發(fā)多臟器功能不全綜合征�,病死率高達(dá)20%~30%,給家庭和社會(huì)帶來沉重負(fù)擔(dān)�。因此,探索SAP的發(fā)病機(jī)制��,尋找有效的治療藥物及方法顯得尤為重要�。
該團(tuán)隊(duì)通過鑒定與紅景天苷相關(guān)的miRNA并探索了其潛在機(jī)制。該團(tuán)隊(duì)通過建立SAP大鼠動(dòng)物模型和miRNA微陣列���,確定紅景天苷對(duì)miRNA表達(dá)譜的影響���,并通過實(shí)時(shí)定量PCR驗(yàn)證它們的變化��。該團(tuán)隊(duì)觀察到����,在這些miRNA中�,經(jīng)紅景天苷處理后miR-217-5p顯著下調(diào)。miR-217-5p過表達(dá)可以通過直接靶向 YY1相關(guān)因子2 (YAF2)的3'UTR來逆轉(zhuǎn)由紅景天苷介導(dǎo)抑制的免疫反應(yīng)����。
該研究揭示了紅景天苷通過調(diào)節(jié)炎癥反應(yīng)發(fā)揮對(duì)SAP的保護(hù)作用,而在調(diào)節(jié)炎癥反應(yīng)的機(jī)制中涉及了miR-217-5p�����。此外����,抑制miR-217-5p可通過靶向YAF2的3'UTR 促進(jìn)YAF2表達(dá),隨后激活p38 MAPK 通路�。(見Figure1, 2, 3, 4)
該研究結(jié)果為紅景天苷治療SAP的作用機(jī)制提出了新的見解,并有助于制定SAP的有效治療策略。
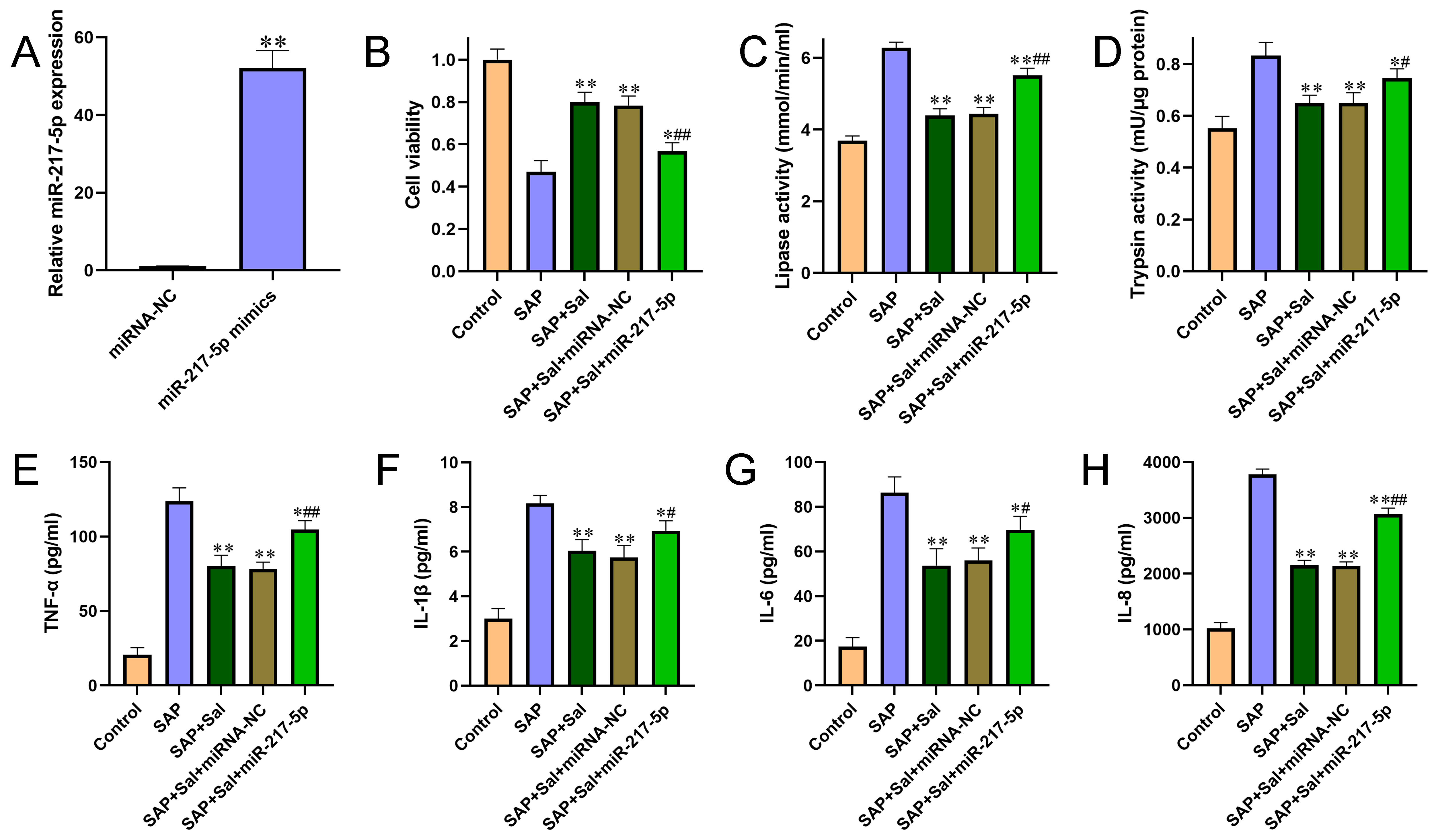
Figure 1 Overexpression of miR-217-5p aggravated cell injury and promoted pro-inflammatory cytokine production. (A) The overexpression efficiency of miR-217-5p mimics was confirmed by qRT-PCR. (B) The effect of miR-217-5p overexpression on cell viability was evaluated by CCK-8 assay. (C-D) The effects of miR-217-5p overexpression on activities of amylase (C) and lipase (D) were detected by automatic biochemical analysis. (E-H) The levels of pro-inflammatory cytokines TNF-α (E), IL? 1β (F), IL-6 (G) and IL-8 (H) from cultured cell medium were determined by ELISA kits according to instructions of the manufacturer. (I-K) Pearson’s correlation analysis was performed to analyze the association between miR-217-5p expression and pancreatic injury score (I), serum amylase activities (J) and serum lipase activities (K). For B-H, *P<0.05, **P<0.01 compared with SAP group, #P<0.05, ##P<0.01 compared with miRNA negative control group.
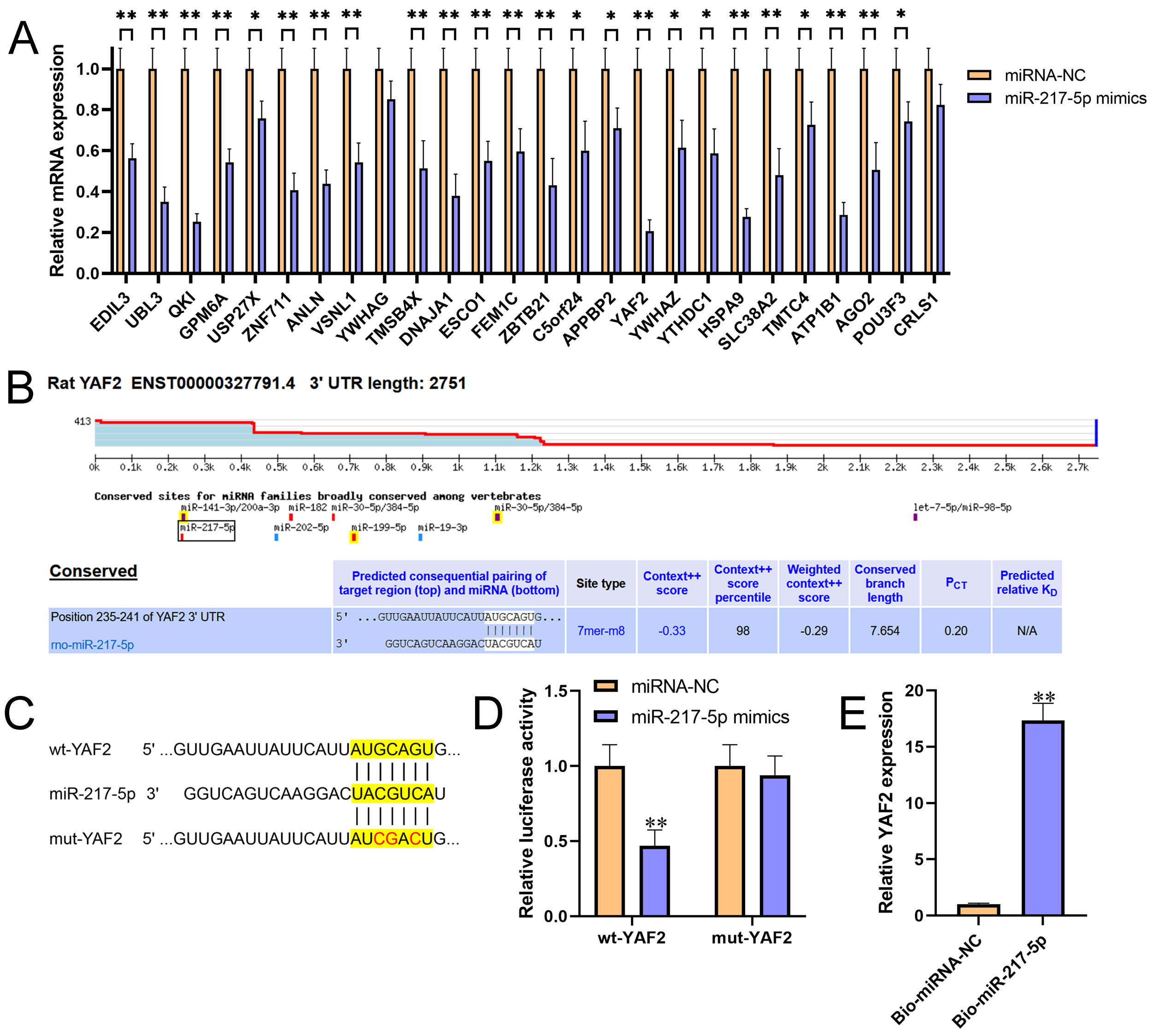
Figure 2 miR-217-5p directly targeted the 3’UTR of YAF2. (A) Relative expression levels of 26 candidate target genes for miR-217-5p were detected by qRT-PCR in AR42J cells after transfected with miR-217-5p mimics or miRNA negative control. (B) The binding position and complementary sequences of miR-217-5p in the 3’UTR of YAF2, which was predicted by TargetScan. (C) The wt- and mut-YAF2 3’UTR sequences for miR-217-5p. (D) Luciferase reporter constructs containing wt- and mut-YAF2 3’UTR were co-transfected with miR-217-5p mimics or miRNA negative control into AR42J cells. After 48h transfection, the luciferase activities were measured. (E) Relative level of YAF2 recruited by Bio-miR-217-5p or Bio-miRNA-NC probes were measured by miRNA pull-down assay. **P<0.01.
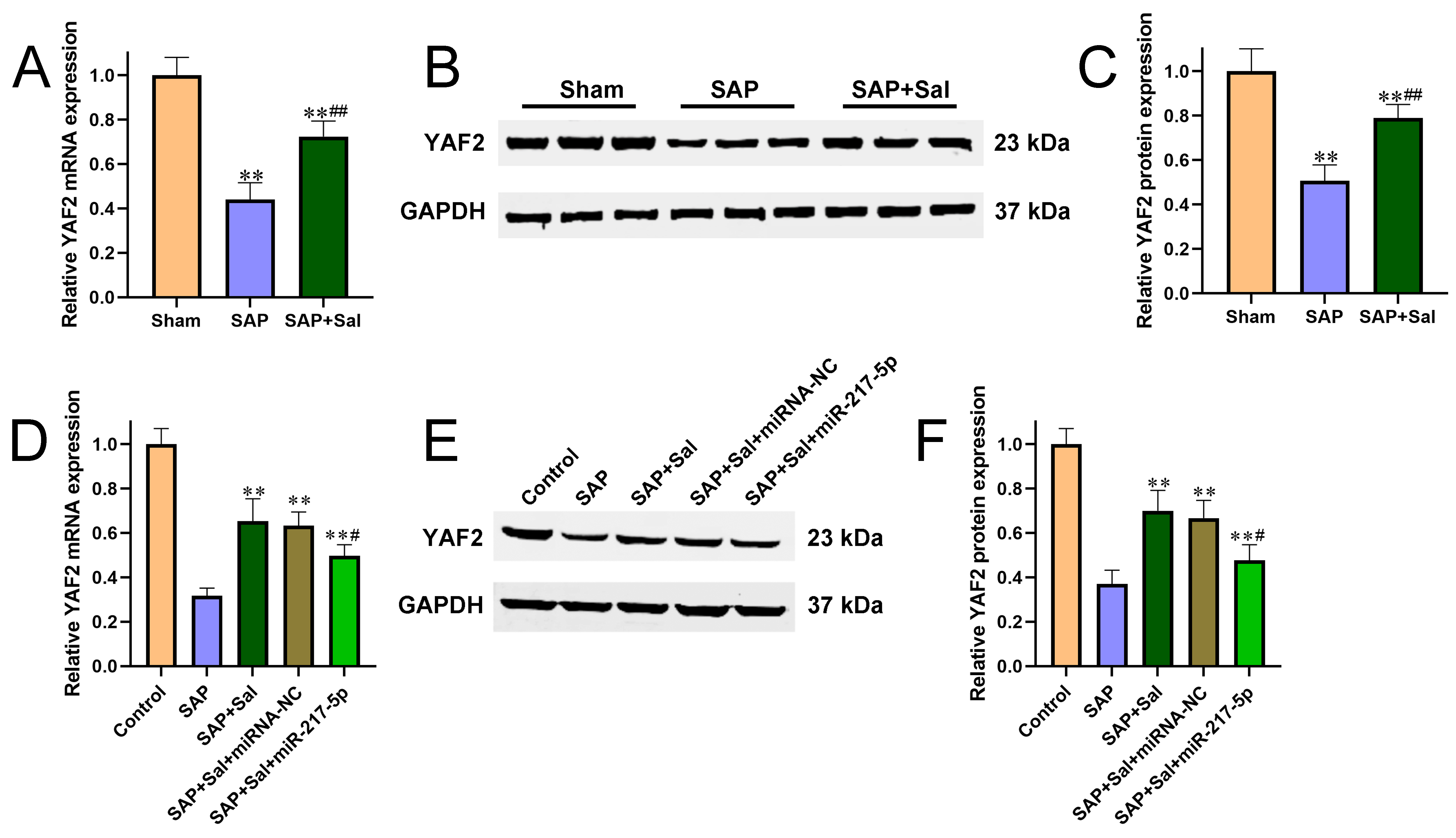
Figure 3 Sal upregulated YAF2 expression via downregulating miR-217-5p. (A-C) The mRNA (A) and protein (B-C) levels of YAF2 in SAP rat pancreatic tissues were determined by qRT-PCR and Western blot, respectively. (D-F) The mRNA (D) and protein (E-F) levels of YAF2 in AR42J cells were determined by qRT-PCR and Western blot, respectively. **P<0.01 compared with sham group, #P<0.05, ##P<0.01 compared with SAP group.
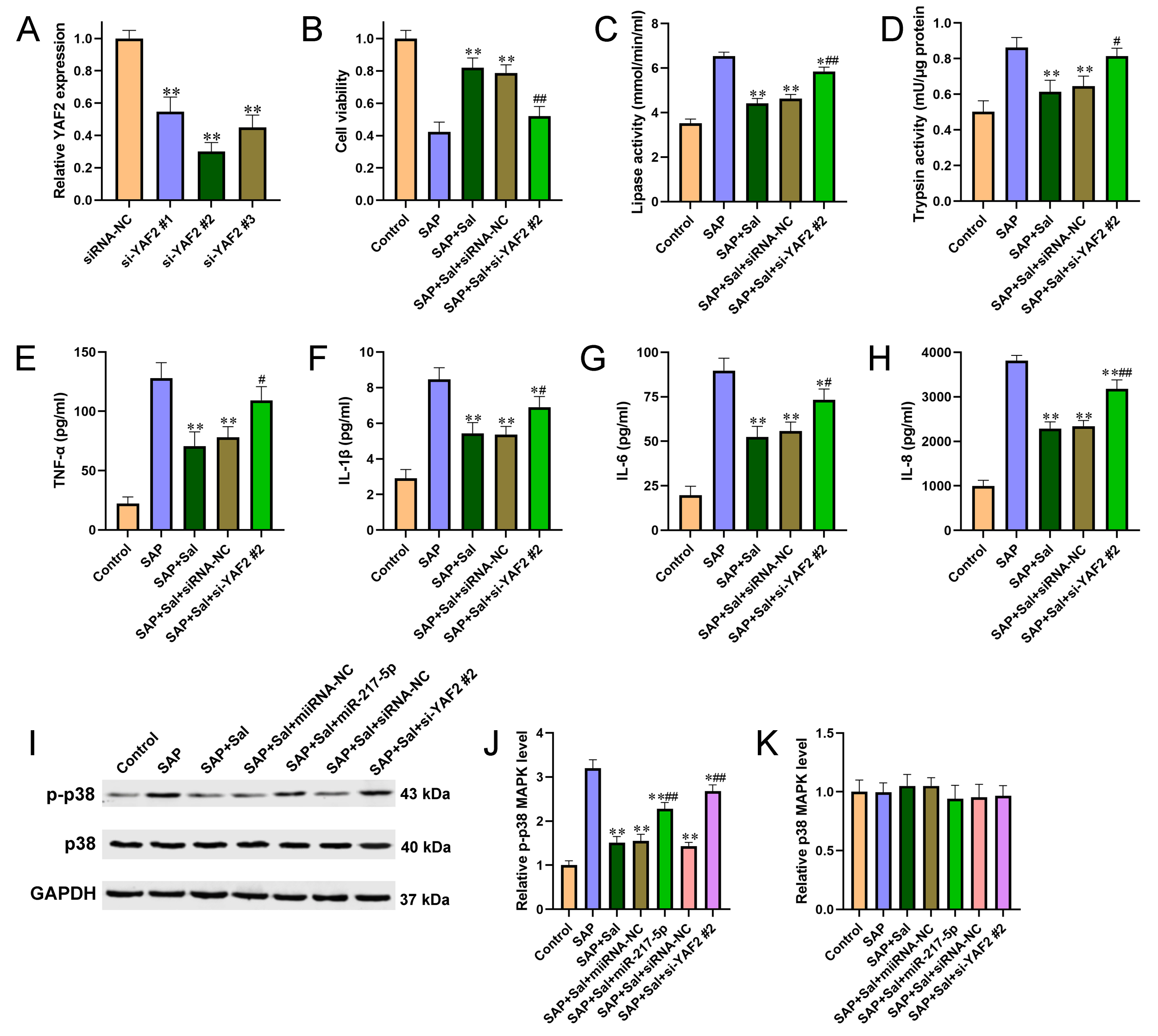
Figure 4 Knockdown of YAF2 reversed Sal induced alleviation of cell injury and inflammation in a YAF2-dependent manner and involving in p38 MAPK pathway. (A) siRNA specific for YAF2 was transfected into AR42J cells and the knockdown efficiency was validated by qRT-PCR. (B) Cell viability was detected by CCK-8 assay. (C-D) The effects of YAF2 knockdown on activities of amylase (C) and lipase (D) were determined by automatic biochemical analysis. (E-H) The effects of YAF2 knockdown on the pro-inflammatory cytokines TNF-α (E), IL? 1β (F), IL-6 (G) and IL-8 (H) were determined by ELISA kits. (I) Western blot was performed to detect the expression levels of p38 and p-p38 in AR42J cells after different treatment. (J-K) Quantitative analysis of p-p38 (J) and p38 (K) from 7I. For A, **P<0.01 compared with siRNA negative control. For B-K, *P<0.05, **P<0.01 compared with SAP group, #P<0.05, ##P<0.01 compared with miRNA or siRNA negative control group.